第 117 章 從流行病學的角度初步分析生存數據 introduction to survival analysis
在本次練習題中,我們學習如何繪製 Kaplan-Meier 生存曲線,並且學會使用 Logrank 檢驗方法來比較不同組之間的生存曲線是否有統計學上的不一致。
117.1 Q1
打開數據 trinmlsh。使用 Stata 的 describe 和 sum 命令熟悉該數據的各個變量的名字和大致內容。主要是確認一下哪些變量含有重要的結果變量信息,和生存時間信息。help trinmlsh 可以調取關於該數據更加詳細的背景和內容介紹。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use trinmlsh
##
## .
## . describe
##
## Contains data from trinmlsh.dta
## obs: 318
## vars: 18 14 Sep 1999 11:20
## -------------------------------------------------------------------------------
## storage display value
## variable name type format label variable label
## -------------------------------------------------------------------------------
## ethgp float %9.0g lethgp Ethnic group
## ageent float %9.0g Age in years at first survey
## death float %9.0g Died from any cause
## cvdeath float %9.0g Died from CV disease
## alc float %9.0g lalc drinks per week
## smokenum float %9.0g lsmokn No. of cigarettes per day
## hdlc float %9.0g HDL cholesterol
## diabp float %9.0g Diastolic blood pressure
## sysbp float %9.0g Systolic blood pressure
## chdstart float %9.0g Heart disease at time of entry
## days float %9.0g
## years float %9.0g
## bmi float %9.0g
## id float %9.0g
## timein float %d
## timeout float %d
## y float %9.0g
## timebth float %d
## -------------------------------------------------------------------------------
## Sorted by:
##
## . sum
##
## Variable | Obs Mean Std. Dev. Min Max
## -------------+---------------------------------------------------------
## ethgp | 318 2.103774 1.347477 1 5
## ageent | 318 65.09343 3.19148 60.005 73.93
## death | 318 .2767296 .4480867 0 1
## cvdeath | 318 .0691824 .254164 0 1
## alc | 318 1.031447 1.111576 0 3
## -------------+---------------------------------------------------------
## smokenum | 317 1.388013 1.649007 0 5
## hdlc | 301 .9880432 .3728258 .185 2.411
## diabp | 314 91.08917 15.12942 55 159
## sysbp | 314 154.8121 28.18775 95 243
## chdstart | 290 .1310345 .3380214 0 1
## -------------+---------------------------------------------------------
## days | 318 2532.101 940.5925 6 3579
## years | 318 6.932514 2.575202 .0164271 9.798768
## bmi | 283 23.25392 4.174109 11.25 36.38
## id | 318 159.5 91.94292 1 318
## timein | 318 6210 0 6210 6210
## -------------+---------------------------------------------------------
## timeout | 318 8742.101 940.5925 6216 9789
## y | 318 6.932514 2.575202 .0164271 9.798768
## timebth | 318 -17565.37 1165.688 -20792.93 -15706.83
##
## .
## .help trinmlshCOHORT STUDY OF RISK FACTORS FOR MORTALITY AMONG MALES IN TRINIDAD
All males aged 35-74 years who were living in two neighbouring suburbs
of Port of Spain, Trinidad, in March 1977 were eligible and entered into
the study. Baseline data were recorded for 1,343 men on a range of risk
factors including ethnic group, blood pressure, glucoose and lipoprotein
concentrations, diabetes mellitus, and cigarette and alchool consumption.
All subjects were then visited annually at home, and morbidity and
mortality records were compiled. Regular inspection of hospital records,
death registers and obitaries were also used to update the records.
Those who had moved away (or abroad) were contacted annually by postal
questionnaire and were also seen if they returned to to Port of Spain.
By these means, loss to follow-up was kept very low.
Follow-up of the study cohort finished at the end of 1986, giving a study
period of almost ten years.
The file trinmlsh contains data on selected risk factors for the
subset of men aged 60 years or over. There were 318 men in this group, and
88 deaths were recorded. Of these deaths, 22 were attributed to
cardiovascular disease. The file holds the following variables:
ethgp ethnic group (1=African,2=Indian,3=European,4=mixed,5=Chin/Sem)
ageent age in years at first survey
death death from any cause (0=no,1=yes)
cvdeath death from CV disease (0=no,1=yes)
alc Drinks per week (0=none,1=1-4,2=5-14,3=15+)
smokenum No. cigs per day (0=non-sm,1=ex,2=1-9,3=10-19,4=20-29,5=30+)
hdlc HDL cholesterol
diabp diastolic BP (mm Hg)
sysbp systolic BP (mm Hg)
chdstart heart disease at time of entry (0=no,1=yes)
days days of follow-up
years years of follow-up
bmi body mass index (wt/(ht*ht))
id Subject identifier
timein Date of entry (days since 1/1/1960)
timeout Date of exit (days since 1/1/1960)
timebth Date of birth (days since 1/1/1960)我們從數據的簡介和變量名字中不難猜出,death 是該研究對象的總死亡結果變量,cvdeath 則特別是死於心血管疾病的結果變量。另外 years 和 y 是內容相同的追蹤時間長度變量,單位是“年”,另一個含有相同信息的時間變量是 days “日”。
117.2 Q2
分析該數據中男性的總死亡 death,繪製總體人羣的一張 Kaplan-Meier 生存曲線圖。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use trinmlsh
##
## .
## . stset timeout, fail(death) origin(timein) enter(timein) scale(365.25) id(id)
##
## id: id
## failure event: death != 0 & death < .
## obs. time interval: (timeout[_n-1], timeout]
## enter on or after: time timein
## exit on or before: failure
## t for analysis: (time-origin)/365.25
## origin: time timein
##
## ------------------------------------------------------------------------------
## 318 total observations
## 0 exclusions
## ------------------------------------------------------------------------------
## 318 observations remaining, representing
## 318 subjects
## 88 failures in single-failure-per-subject data
## 2,204.539 total analysis time at risk and under observation
## at risk from t = 0
## earliest observed entry t = 0
## last observed exit t = 9.798768
##
## .在 Stata 繪製 Kaplan-Meier 生存曲線極其簡單:
sts graph, saving(plot1)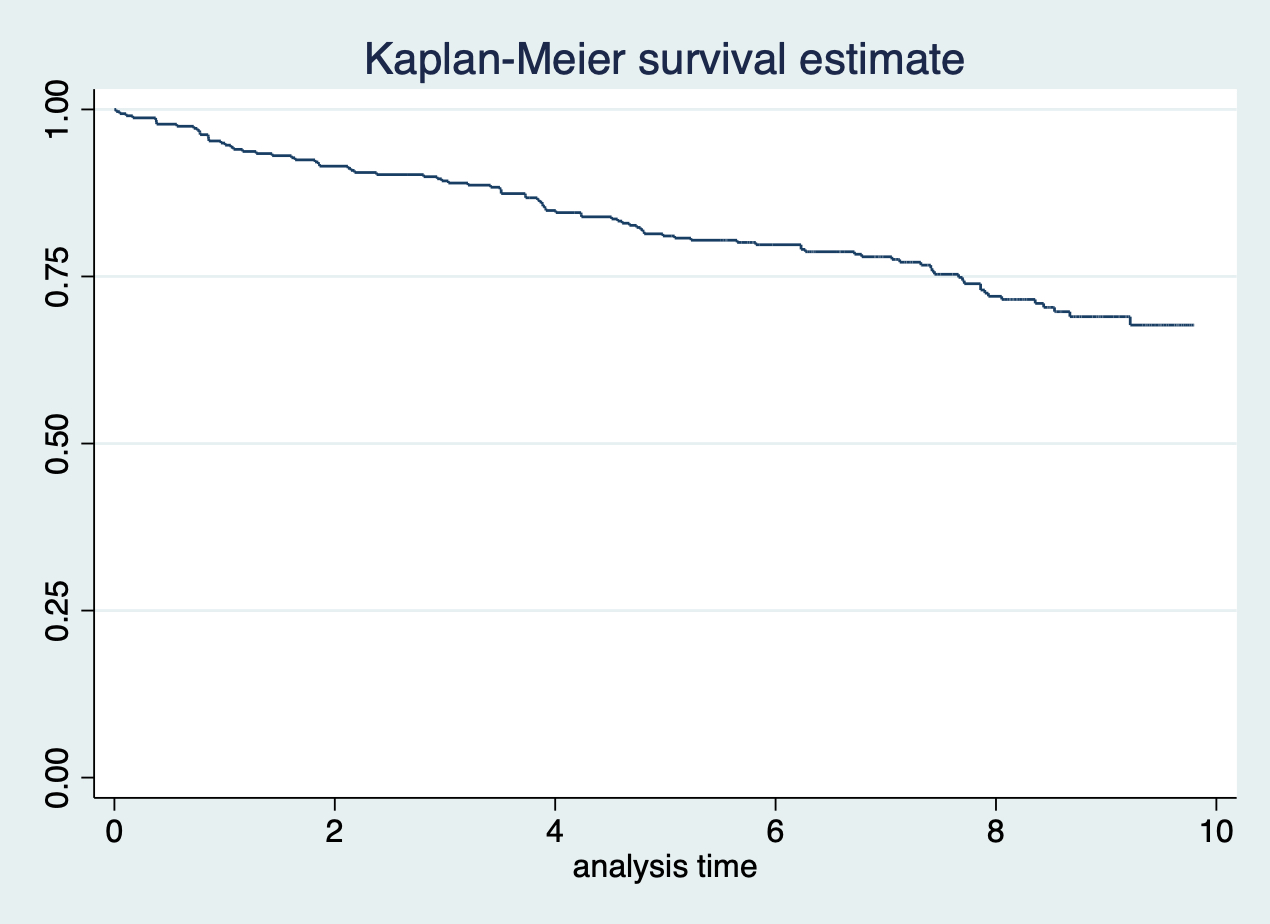
圖 117.1: sts graph, saving(plot1)
117.3 Q3
使用 sts list 命令來查看該數據中第1,3,和5年時的累計生存概率 (cumulative survival probability)。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use trinmlsh
##
## . quietly stset timeout, fail(death) origin(timein) enter(timein) scale(365.25)
## > id(id)
##
## . sts list, at(1, 3, 5)
##
## failure _d: death
## analysis time _t: (timeout-origin)/365.25
## origin: time timein
## enter on or after: time timein
## id: id
##
## Beg. Survivor Std.
## Time Total Fail Function Error [95% Conf. Int.]
## -------------------------------------------------------------------------------
## 1 303 16 0.9497 0.0123 0.9192 0.9689
## 3 284 18 0.8930 0.0173 0.8536 0.9224
## 5 256 26 0.8107 0.0220 0.7630 0.8497
## -------------------------------------------------------------------------------
## Note: Survivor function is calculated over full data and evaluated at indicated
## times; it is not calculated from aggregates shown at left.
##
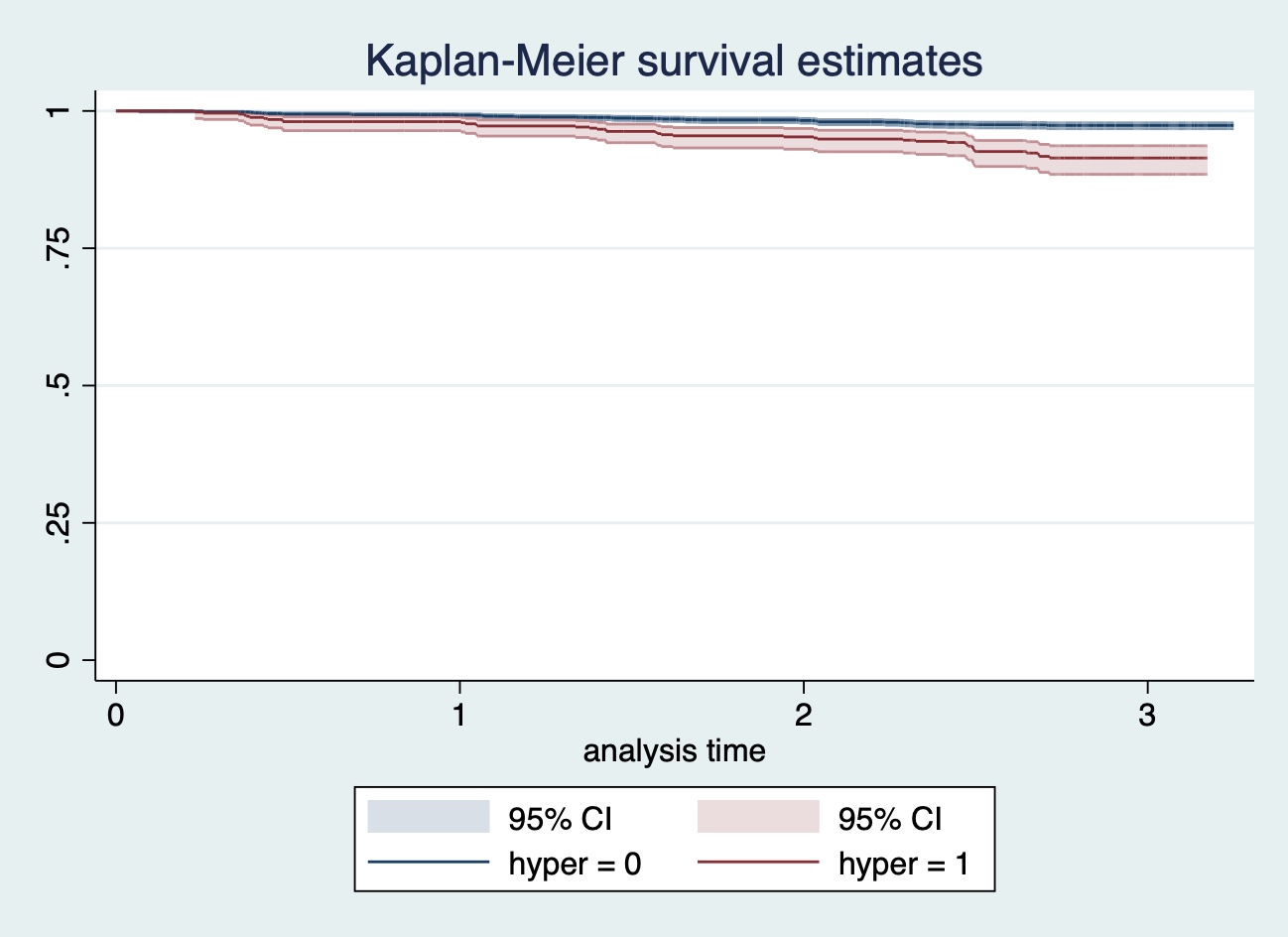
## .117.4 Q4
給Q2 的 Kaplan-Meier 曲線添加 95% 信賴區間:
sts graph, ci saving(plot1, replace)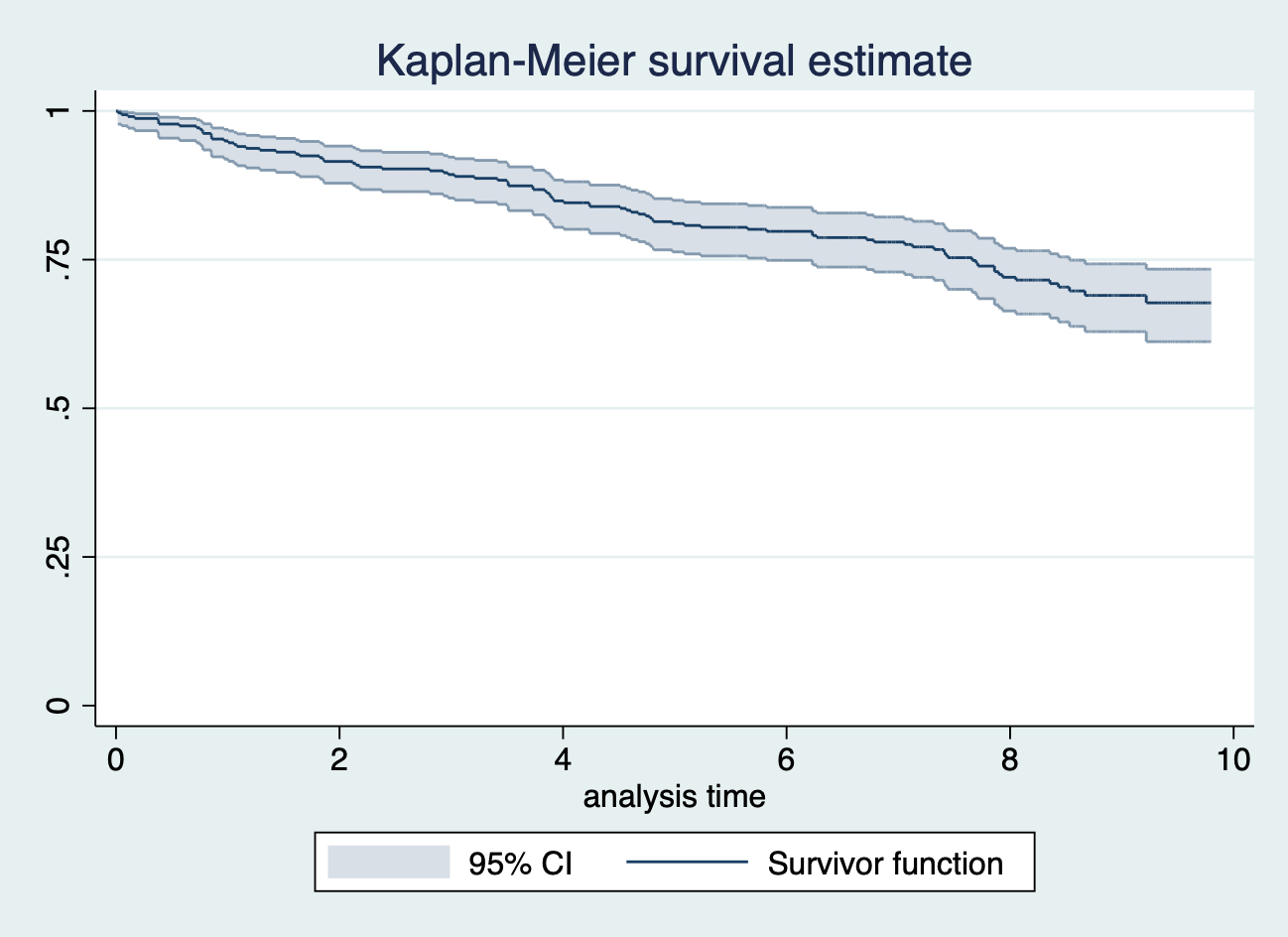
圖 117.2: sts graph, ci saving(plot1, replace)
117.5 Q5
關注變量 smokenum, 查看它的詳細內容。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use trinmlsh
##
## . quietly stset timeout, fail(death) origin(timein) enter(timein) scale(365.25)
## > id(id)
##
## . tab smokenum
##
## No. of |
## cigarettes |
## per day | Freq. Percent Cum.
## ------------+-----------------------------------
## non-smok | 140 44.16 44.16
## ex-smoke | 68 21.45 65.62
## 1-9 cigs | 29 9.15 74.76
## 10-19 ci | 30 9.46 84.23
## 20-29 ci | 26 8.20 92.43
## 30+ cigs | 24 7.57 100.00
## ------------+-----------------------------------
## Total | 317 100.00
##
## . tab smokenum, nolabel
##
## No. of |
## cigarettes |
## per day | Freq. Percent Cum.
## ------------+-----------------------------------
## 0 | 140 44.16 44.16
## 1 | 68 21.45 65.62
## 2 | 29 9.15 74.76
## 3 | 30 9.46 84.23
## 4 | 26 8.20 92.43
## 5 | 24 7.57 100.00
## ------------+-----------------------------------
## Total | 317 100.00
##
## .生成一個新的變量區分對象是否是現在有吸菸習慣 “current smoker”。比較現在有吸菸習慣和無吸菸習慣的兩組對象的生存曲線,這兩條曲線之間是否有(統計學上的)不同?
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use trinmlsh
##
## . quietly stset timeout, fail(death) origin(timein) enter(timein) scale(365.25)
## > id(id)
##
## . gen smokstatus = smokenum >= 2 if smokenum != .
## (1 missing value generated)
##
## . label define smokstatus 0 "non-smokers" 1 "current smokers"
##
## . label value smokstatus smokstatus
##
## .繪製兩組人的生存概率曲線,需要用到 by(smokstatus) 選項。
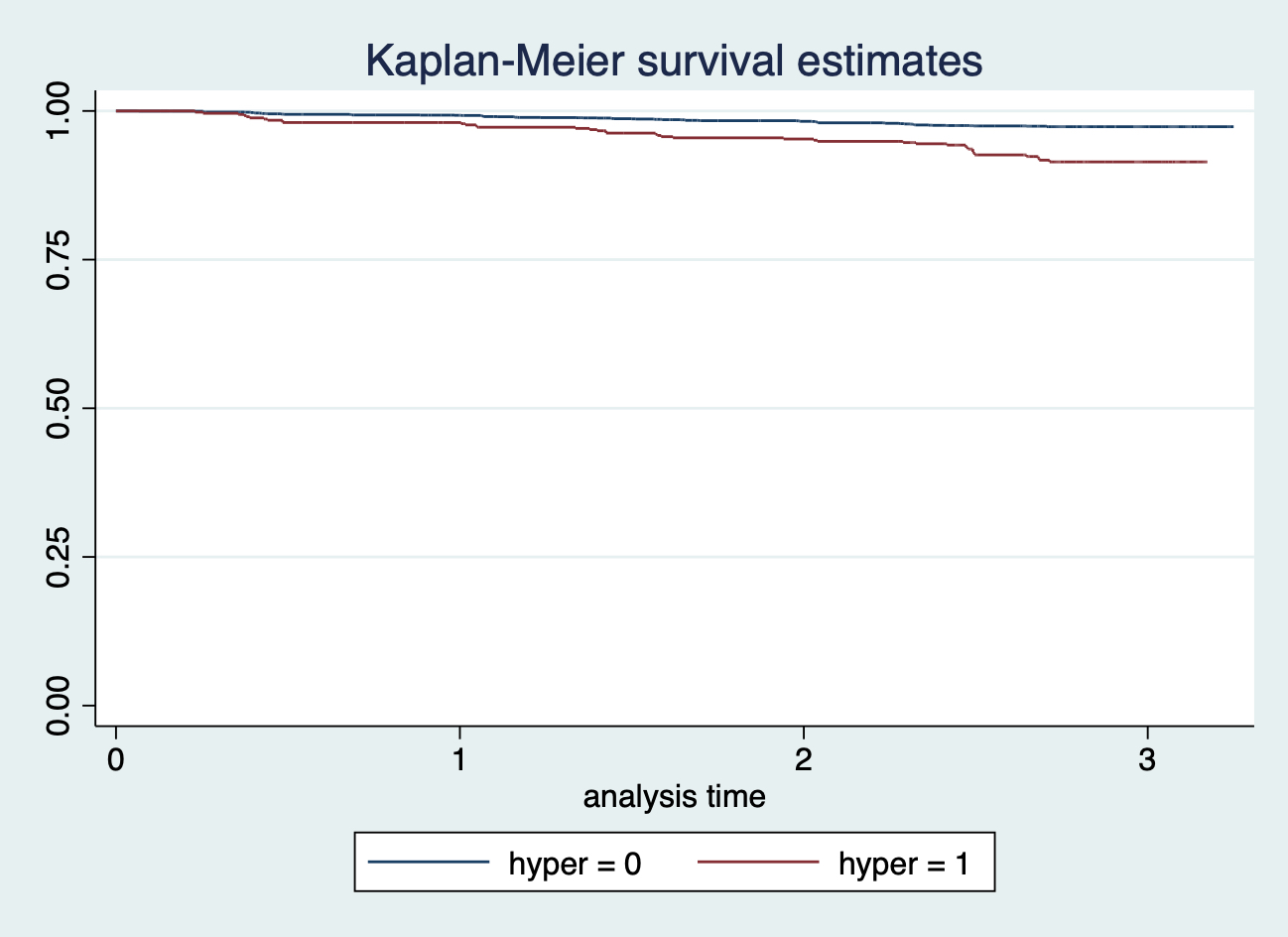
sts graph, by(smokstatus)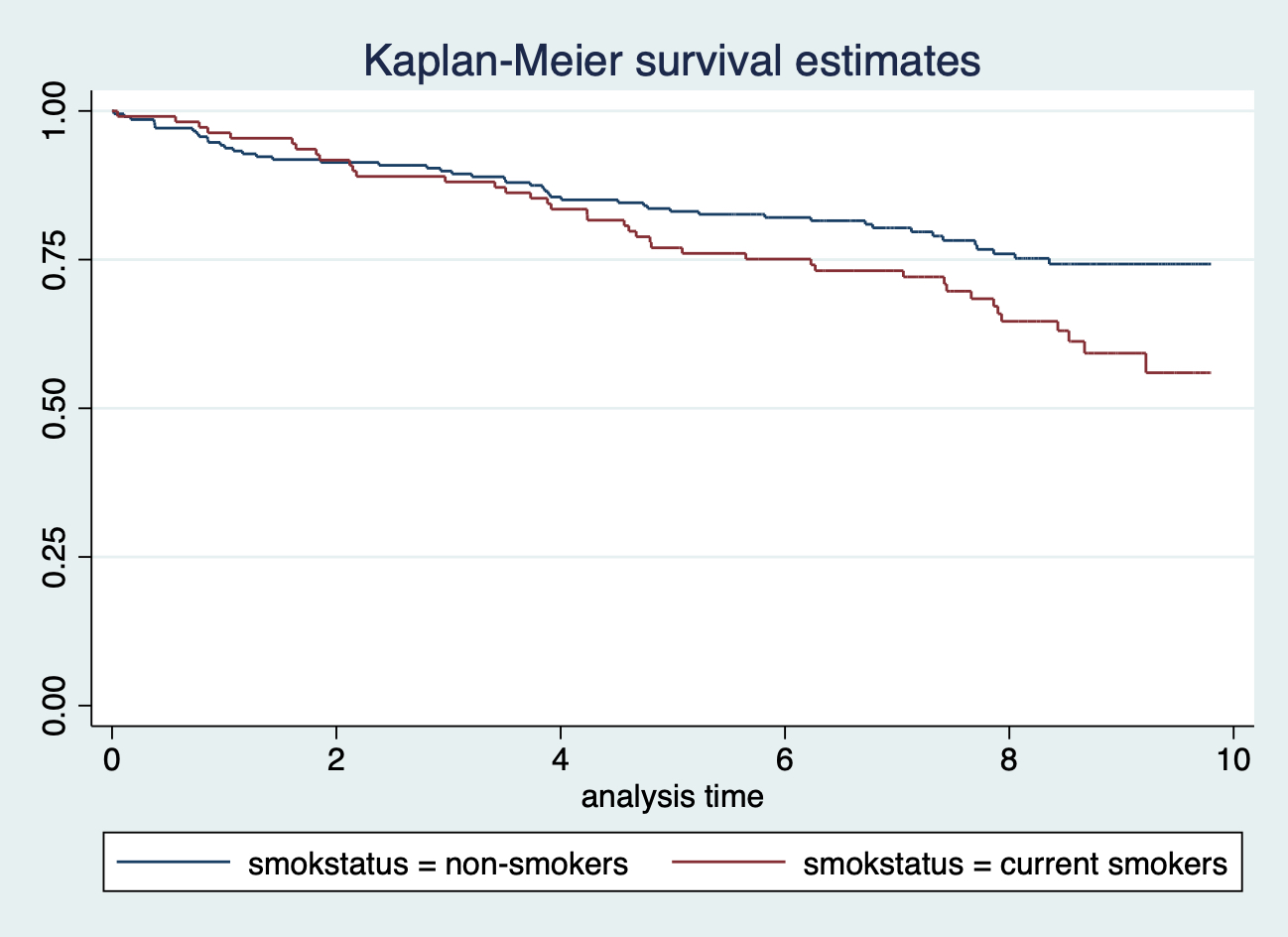
圖 117.3: sts graph, by(smokstatus)
可以看到吸菸和不吸菸兩組人的生存曲線在追蹤到第四年左右之前都十分相似。但是再往後的觀察隨訪時間裏,吸菸者的希望概率似乎要更高。
117.6 Q6
檢驗吸菸習慣不同的兩組實驗對象的生存曲線在整個隨訪過程中是否有統計學意義上的不同?
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use trinmlsh
##
## . quietly stset timeout, fail(death) origin(timein) enter(timein) scale(365.25)
## > id(id)
##
## . gen smokstatus = smokenum >= 2 if smokenum != .
## (1 missing value generated)
##
## . label define smokstatus 0 "non-smokers" 1 "current smokers"
##
## . label value smokstatus smokstatus
##
## . sts test smokstatus
##
## failure _d: death
## analysis time _t: (timeout-origin)/365.25
## origin: time timein
## enter on or after: time timein
## id: id
##
##
## Log-rank test for equality of survivor functions
## ------------------------------------------------
##
## | Events Events
## smokstatus | observed expected
## ----------------+-------------------------
## non-smokers | 48 57.86
## current smokers | 40 30.14
## ----------------+-------------------------
## Total | 88 88.00
##
## chi2(1) = 4.91
## Pr>chi2 = 0.0267
##
## .Logrank 檢驗結果提示吸菸和不吸菸兩組人的生存曲線確實存在統計學上的差異 (p = 0.03)。
117.7 Q7
打開 mortality 數據。該隊列研究在北部尼日利亞地區實施了三年。我們主要在這裏觀察分析高血壓和總死亡之間的關係。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . describe
##
## Contains data from mortality.dta
## obs: 4,298
## vars: 28 23 Oct 2013 11:45
## -------------------------------------------------------------------------------
## storage display value
## variable name type format label variable label
## -------------------------------------------------------------------------------
## id int %8.0g Individual ID
## area float %9.0g area Area
## district str7 %7s District
## vcode byte %8.0g Village ID
## compound_size float %9.0g Compound size
## age byte %8.0g Age in years
## sex byte %8.0g sex Sex
## ethnic byte %8.0g ethnic Ethnic
## religion byte %11.0g religion Religion
## occupation byte %27.0g occupation
## Occupation
## education byte %22.0g education
## Education
## mfpermg float %9.0g Microfilarial load per mg
## vimp float %17.0g vimp Visually impaired
## died float %9.0g Died
## systolic int %8.0g Systolic BP (mmHg)
## diastolic int %8.0g Diastolic BP (mmHg)
## pulse int %8.0g Pulse rate/minute
## weight int %8.0g Weight (kgs)
## height int %8.0g Height (cms)
## map float %9.0g Mean Arterial Pressure
## bmi float %9.0g BMI
## agegrp float %9.0g agegrp Age group (years)
## mfgrp float %10.0g mfgrp Microfilarial load/mg (grouped)
## mfpos float %9.0g Microfilarial infection
## agebin float %9.0g agebin Age (2 groups)
## bmigrp float %12.0g bmigrp BMI (3 groups)
## enter long %dD_m_Y Date of entry to study
## exit float %dD_m_Y Date of exit from study
## -------------------------------------------------------------------------------
## Sorted by: id
##
## .117.8 Q8
使用 gen 命令生成一個新的變量,根據收縮期血壓 (based on systolic blood pressure) 定義實驗對象是否有高血壓。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . gen hyper = .
## (4,298 missing values generated)
##
## . replace hyper = 0 if systolic < 140
## (3,747 real changes made)
##
## . replace hyper = 1 if systolic >= 140 & systolic != .
## (516 real changes made)
##
## .117.9 Q9
使用 stset 命令準備你的數據,讓它可以在 Stata 裏實施生存分析。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . stset exit, failure(died) enter(enter) origin(enter) id(id) scale(365.25)
##
## id: id
## failure event: died != 0 & died < .
## obs. time interval: (exit[_n-1], exit]
## enter on or after: time enter
## exit on or before: failure
## t for analysis: (time-origin)/365.25
## origin: time enter
##
## ------------------------------------------------------------------------------
## 4,298 total observations
## 0 exclusions
## ------------------------------------------------------------------------------
## 4,298 observations remaining, representing
## 4,298 subjects
## 137 failures in single-failure-per-subject data
## 11,457.357 total analysis time at risk and under observation
## at risk from t = 0
## earliest observed entry t = 0
## last observed exit t = 3.249829
##
## .117.10 Q10
使用 strate 命令計算有高血壓和無高血壓兩組人個字的總死亡率 all-cause mortality rate。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . gen hyper = .
## (4,298 missing values generated)
##
## . quietly replace hyper = 0 if systolic < 140
##
## . quietly replace hyper = 1 if systolic >= 140 & systolic != .
##
## . quietly stset exit, failure(died) enter(enter) origin(enter) id(id) scale(365
## > .25)
##
## . strate hyper, per(1000)
##
## failure _d: died
## analysis time _t: (exit-origin)/365.25
## origin: time enter
## enter on or after: time enter
## id: id
##
## Estimated failure rates
## Number of records = 4263
##
## +---------------------------------------------------+
## | hyper D Y Rate Lower Upper |
## |---------------------------------------------------|
## | 0 94 9.9879 9.4114 7.6888 11.5199 |
## | 1 40 1.3819 28.9456 21.2322 39.4611 |
## +---------------------------------------------------+
## Notes: Rate = D/Y = failures/person-time (per 1000).
## Lower and Upper are bounds of 95% confidence intervals.
##
##
## .計算結果是高血壓的實驗對象中,總死亡率達到每 1000 人年 28.95,沒有高血壓的人羣的死亡率是9.4 每 1000人年。
117.11 Q11
使用 stmh 命令計算高血壓和總死亡之間的關係,比較有高血壓的人相對無高血壓人的總體死亡率比 rate ratio。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . gen hyper = .
## (4,298 missing values generated)
##
## . quietly replace hyper = 0 if systolic < 140
##
## . quietly replace hyper = 1 if systolic >= 140 & systolic != .
##
## . quietly stset exit, failure(died) enter(enter) origin(enter) id(id) scale(365
## > .25)
##
## . stmh hyper
##
## failure _d: died
## analysis time _t: (exit-origin)/365.25
## origin: time enter
## enter on or after: time enter
## id: id
##
## Mantel-Haenszel estimate of the rate ratio
## comparing hyper==1 vs. hyper==0
##
## ------------------------------------------------------------------------
## Rate ratio chi2 P>chi2 [95% Conf. Interval]
## ------------------------------------------------------------------------
## 3.076 39.30 0.0000 2.124 4.453
## ------------------------------------------------------------------------
##
## .計算死亡率比的結果表示,有高血壓的實驗對象色死亡率是無高血壓的人死亡率的三倍還多。
117.12 Q12-13
這時候,我們使用 browse 瀏覽正在分析的數據時,會發現最後幾個新增的變量。
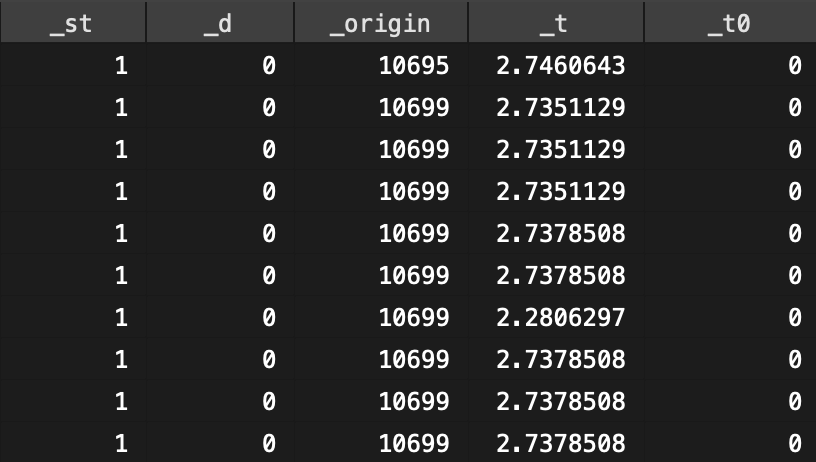
圖 117.4: New variables generated by Stata to calcuate rate.(1)
圖 117.5: New variables generated by Stata to calcuate rate.(2)
新增的幾個變量分別定義爲:
_st is an indicator variable taking the value 0 or 1. 1 indicates records included in the analyses, 0 indicates records excluded from analyses, for example if the exit time was earlier than the entry time
_d records whether the individual experienced the event (=1) or not (=0). In this simple situation _d = died (try tab _d died)
_t is the time the person exited the study (in years since entry to the study)
_t0 is the time the person entered the study (in years since entry to the study, so zero for everyone)117.13 Q14
製作一個生存表格,只保留高血壓人羣。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . gen hyper = .
## (4,298 missing values generated)
##
## . quietly replace hyper = 0 if systolic < 140
##
## . quietly replace hyper = 1 if systolic >= 140 & systolic != .
##
## . quietly stset exit, failure(died) enter(enter) origin(enter) id(id) scale(365
## > .25)
##
## . sts list if hyper == 1
##
## failure _d: died
## analysis time _t: (exit-origin)/365.25
## origin: time enter
## enter on or after: time enter
## id: id
##
## At Net Survivor Std.
## Time Risk Fail Lost Function Error [95% Conf. Int.]
## ------------------------------------------------------------------------
## .23 516 1 0 0.9981 0.0019 0.9863 0.9997
## .2533 515 1 0 0.9961 0.0027 0.9846 0.9990
## .3559 514 1 0 0.9942 0.0033 0.9821 0.9981
## .3587 513 0 1 0.9942 0.0033 0.9821 0.9981
## .3669 512 0 1 0.9942 0.0033 0.9821 0.9981
## .3723 511 1 0 0.9922 0.0039 0.9795 0.9971
## .3792 510 1 0 0.9903 0.0043 0.9768 0.9959
## .3888 509 1 0 0.9883 0.0047 0.9743 0.9947
## .4298 508 1 0 0.9864 0.0051 0.9717 0.9935
## .4353 507 0 1 0.9864 0.0051 0.9717 0.9935
## .4408 506 1 0 0.9845 0.0055 0.9692 0.9922
## .4846 505 2 0 0.9806 0.0061 0.9642 0.9895
## .5421 503 0 1 0.9806 0.0061 0.9642 0.9895
## .6886 502 0 1 0.9806 0.0061 0.9642 0.9895
## .8378 501 0 1 0.9806 0.0061 0.9642 0.9895
## .9363 500 0 1 0.9806 0.0061 0.9642 0.9895
## .9514 499 0 1 0.9806 0.0061 0.9642 0.9895
## 1.003 498 1 0 0.9786 0.0064 0.9617 0.9881
## 1.012 497 0 1 0.9786 0.0064 0.9617 0.9881
## 1.016 496 1 0 0.9766 0.0067 0.9592 0.9867
## 1.047 495 1 0 0.9746 0.0069 0.9567 0.9852
## 1.05 494 1 0 0.9727 0.0072 0.9543 0.9837
## 1.055 493 0 1 0.9727 0.0072 0.9543 0.9837
## 1.336 492 1 0 0.9707 0.0075 0.9519 0.9822
## 1.377 491 1 0 0.9687 0.0077 0.9494 0.9807
## 1.399 490 1 0 0.9667 0.0079 0.9470 0.9792
## 1.418 489 0 1 0.9667 0.0079 0.9470 0.9792
## 1.425 488 2 0 0.9628 0.0084 0.9423 0.9761
## 1.477 486 0 1 0.9628 0.0084 0.9423 0.9761
## 1.569 485 1 0 0.9608 0.0086 0.9399 0.9745
## 1.574 484 1 0 0.9588 0.0088 0.9375 0.9729
## 1.585 483 1 0 0.9568 0.0090 0.9352 0.9714
## 1.619 482 1 0 0.9548 0.0092 0.9328 0.9698
## 1.941 481 1 1 0.9528 0.0094 0.9305 0.9681
## 2.029 479 1 0 0.9509 0.0096 0.9281 0.9665
## 2.04 478 1 0 0.9489 0.0098 0.9258 0.9649
## 2.277 477 0 1 0.9489 0.0098 0.9258 0.9649
## 2.286 476 0 1 0.9489 0.0098 0.9258 0.9649
## 2.289 475 1 0 0.9469 0.0100 0.9235 0.9633
## 2.311 474 0 2 0.9469 0.0100 0.9235 0.9633
## 2.313 472 0 3 0.9469 0.0100 0.9235 0.9633
## 2.316 469 0 2 0.9469 0.0100 0.9235 0.9633
## 2.319 467 0 5 0.9469 0.0100 0.9235 0.9633
## 2.322 462 0 2 0.9469 0.0100 0.9235 0.9633
## 2.326 460 1 0 0.9448 0.0101 0.9211 0.9616
## 2.404 459 0 1 0.9448 0.0101 0.9211 0.9616
## 2.407 458 0 2 0.9448 0.0101 0.9211 0.9616
## 2.409 456 0 3 0.9448 0.0101 0.9211 0.9616
## 2.412 453 0 7 0.9448 0.0101 0.9211 0.9616
## 2.415 446 0 4 0.9448 0.0101 0.9211 0.9616
## 2.416 442 1 0 0.9427 0.0103 0.9185 0.9598
## 2.439 441 0 3 0.9427 0.0103 0.9185 0.9598
## 2.442 438 0 2 0.9427 0.0103 0.9185 0.9598
## 2.445 436 0 5 0.9427 0.0103 0.9185 0.9598
## 2.448 431 0 3 0.9427 0.0103 0.9185 0.9598
## 2.45 428 0 2 0.9427 0.0103 0.9185 0.9598
## 2.453 426 0 5 0.9427 0.0103 0.9185 0.9598
## 2.471 421 1 0 0.9404 0.0106 0.9159 0.9580
## 2.472 420 1 0 0.9382 0.0108 0.9132 0.9561
## 2.478 419 1 0 0.9360 0.0110 0.9106 0.9543
## 2.48 418 0 4 0.9360 0.0110 0.9106 0.9543
## 2.483 414 0 3 0.9360 0.0110 0.9106 0.9543
## 2.486 411 0 9 0.9360 0.0110 0.9106 0.9543
## 2.489 402 0 14 0.9360 0.0110 0.9106 0.9543
## 2.49 388 0 1 0.9360 0.0110 0.9106 0.9543
## 2.491 387 2 8 0.9311 0.0114 0.9048 0.9503
## 2.494 377 1 7 0.9287 0.0117 0.9019 0.9483
## 2.497 369 1 3 0.9261 0.0119 0.8989 0.9462
## 2.5 365 0 5 0.9261 0.0119 0.8989 0.9462
## 2.502 360 0 2 0.9261 0.0119 0.8989 0.9462
## 2.505 358 0 3 0.9261 0.0119 0.8989 0.9462
## 2.639 355 0 2 0.9261 0.0119 0.8989 0.9462
## 2.642 353 0 8 0.9261 0.0119 0.8989 0.9462
## 2.645 345 0 8 0.9261 0.0119 0.8989 0.9462
## 2.648 337 1 7 0.9234 0.0122 0.8956 0.9440
## 2.65 329 0 6 0.9234 0.0122 0.8956 0.9440
## 2.653 323 0 6 0.9234 0.0122 0.8956 0.9440
## 2.656 317 0 8 0.9234 0.0122 0.8956 0.9440
## 2.667 309 0 1 0.9234 0.0122 0.8956 0.9440
## 2.68 308 0 1 0.9234 0.0122 0.8956 0.9440
## 2.682 307 1 1 0.9204 0.0125 0.8919 0.9416
## 2.684 305 1 0 0.9174 0.0128 0.8882 0.9392
## 2.715 304 1 0 0.9143 0.0131 0.8846 0.9367
## 2.719 303 0 1 0.9143 0.0131 0.8846 0.9367
## 2.721 302 0 1 0.9143 0.0131 0.8846 0.9367
## 2.727 301 0 3 0.9143 0.0131 0.8846 0.9367
## 2.73 298 0 14 0.9143 0.0131 0.8846 0.9367
## 2.732 284 0 6 0.9143 0.0131 0.8846 0.9367
## 2.735 278 0 4 0.9143 0.0131 0.8846 0.9367
## 2.738 274 0 4 0.9143 0.0131 0.8846 0.9367
## 2.741 270 0 13 0.9143 0.0131 0.8846 0.9367
## 2.743 257 0 4 0.9143 0.0131 0.8846 0.9367
## 2.746 253 0 5 0.9143 0.0131 0.8846 0.9367
## 2.757 248 0 1 0.9143 0.0131 0.8846 0.9367
## 2.793 247 0 1 0.9143 0.0131 0.8846 0.9367
## 2.812 246 0 1 0.9143 0.0131 0.8846 0.9367
## 2.815 245 0 7 0.9143 0.0131 0.8846 0.9367
## 2.817 238 0 3 0.9143 0.0131 0.8846 0.9367
## 2.823 235 0 2 0.9143 0.0131 0.8846 0.9367
## 2.825 233 0 4 0.9143 0.0131 0.8846 0.9367
## 2.828 229 0 4 0.9143 0.0131 0.8846 0.9367
## 2.831 225 0 3 0.9143 0.0131 0.8846 0.9367
## 2.834 222 0 1 0.9143 0.0131 0.8846 0.9367
## 2.875 221 0 2 0.9143 0.0131 0.8846 0.9367
## 2.899 219 0 1 0.9143 0.0131 0.8846 0.9367
## 2.962 218 0 1 0.9143 0.0131 0.8846 0.9367
## 2.965 217 0 8 0.9143 0.0131 0.8846 0.9367
## 2.968 209 0 7 0.9143 0.0131 0.8846 0.9367
## 2.971 202 0 9 0.9143 0.0131 0.8846 0.9367
## 2.973 193 0 20 0.9143 0.0131 0.8846 0.9367
## 2.976 173 0 8 0.9143 0.0131 0.8846 0.9367
## 2.979 165 0 5 0.9143 0.0131 0.8846 0.9367
## 2.982 160 0 3 0.9143 0.0131 0.8846 0.9367
## 2.984 157 0 3 0.9143 0.0131 0.8846 0.9367
## 2.99 154 0 2 0.9143 0.0131 0.8846 0.9367
## 2.995 152 0 1 0.9143 0.0131 0.8846 0.9367
## 3.001 151 0 1 0.9143 0.0131 0.8846 0.9367
## 3.039 150 0 4 0.9143 0.0131 0.8846 0.9367
## 3.042 146 0 2 0.9143 0.0131 0.8846 0.9367
## 3.044 144 0 3 0.9143 0.0131 0.8846 0.9367
## 3.047 141 0 4 0.9143 0.0131 0.8846 0.9367
## 3.05 137 0 2 0.9143 0.0131 0.8846 0.9367
## 3.053 135 0 3 0.9143 0.0131 0.8846 0.9367
## 3.061 132 0 1 0.9143 0.0131 0.8846 0.9367
## 3.072 131 0 2 0.9143 0.0131 0.8846 0.9367
## 3.08 129 0 7 0.9143 0.0131 0.8846 0.9367
## 3.083 122 0 4 0.9143 0.0131 0.8846 0.9367
## 3.086 118 0 3 0.9143 0.0131 0.8846 0.9367
## 3.088 115 0 3 0.9143 0.0131 0.8846 0.9367
## 3.091 112 0 1 0.9143 0.0131 0.8846 0.9367
## 3.113 111 0 1 0.9143 0.0131 0.8846 0.9367
## 3.116 110 0 6 0.9143 0.0131 0.8846 0.9367
## 3.118 104 0 12 0.9143 0.0131 0.8846 0.9367
## 3.121 92 0 8 0.9143 0.0131 0.8846 0.9367
## 3.143 84 0 1 0.9143 0.0131 0.8846 0.9367
## 3.146 83 0 8 0.9143 0.0131 0.8846 0.9367
## 3.149 75 0 13 0.9143 0.0131 0.8846 0.9367
## 3.151 62 0 19 0.9143 0.0131 0.8846 0.9367
## 3.157 43 0 1 0.9143 0.0131 0.8846 0.9367
## 3.159 42 0 4 0.9143 0.0131 0.8846 0.9367
## 3.162 38 0 8 0.9143 0.0131 0.8846 0.9367
## 3.165 30 0 15 0.9143 0.0131 0.8846 0.9367
## 3.168 15 0 10 0.9143 0.0131 0.8846 0.9367
## 3.17 5 0 4 0.9143 0.0131 0.8846 0.9367
## ------------------------------------------------------------------------
## Note: Net Lost equals the number lost minus the number who entered.
##
## .117.16 Q17
Kaplan Meier 生存曲線繪製的是累計生存概率,我們再來嘗試繪製累計死亡率,
sts graph, by(hyper) ci failure ylabel(0 0.05 0.10 0.15 0.20)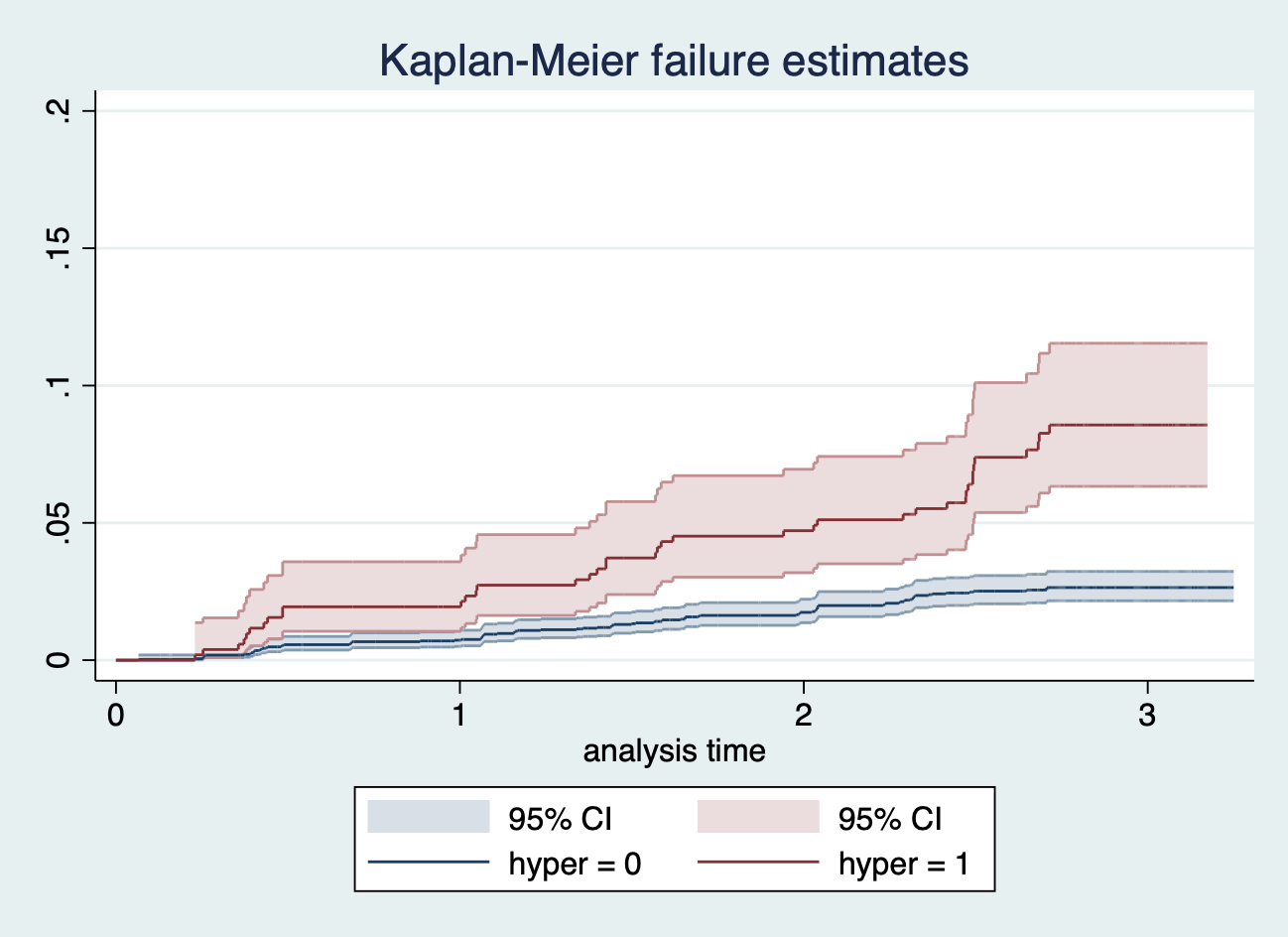
圖 117.8: sts graph, by(hyper) ci failure ylabel(0 0.05 0.10 0.15 0.20)
117.17 Q18
使用 Logrank 檢驗法檢驗有無高血壓之間是否存在統計學上的生存曲線差異。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . gen hyper = .
## (4,298 missing values generated)
##
## . quietly replace hyper = 0 if systolic < 140
##
## . quietly replace hyper = 1 if systolic >= 140 & systolic != .
##
## . quietly stset exit, failure(died) enter(enter) origin(enter) id(id) scale(365
## > .25)
##
## . sts test hyper
##
## failure _d: died
## analysis time _t: (exit-origin)/365.25
## origin: time enter
## enter on or after: time enter
## id: id
##
##
## Log-rank test for equality of survivor functions
## ------------------------------------------------
##
## | Events Events
## hyper | observed expected
## ------+-------------------------
## 0 | 94 117.91
## 1 | 40 16.09
## ------+-------------------------
## Total | 134 134.00
##
## chi2(1) = 40.36
## Pr>chi2 = 0.0000
##
## .我們發現數據提供了極強的證據證明有無高血壓人羣之間確實是存在生存曲線的統計學差異。
117.18 Q19
這次我們重新使用 stset 命令來準備生存數據,但是我們不指定時間的起點 origin:
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . stset exit, failure(died) enter(enter) id(id) scale(365.25)
##
## id: id
## failure event: died != 0 & died < .
## obs. time interval: (exit[_n-1], exit]
## enter on or after: time enter
## exit on or before: failure
## t for analysis: time/365.25
##
## ------------------------------------------------------------------------------
## 4,298 total observations
## 0 exclusions
## ------------------------------------------------------------------------------
## 4,298 observations remaining, representing
## 4,298 subjects
## 137 failures in single-failure-per-subject data
## 11,457.357 total analysis time at risk and under observation
## at risk from t = 0
## earliest observed entry t = 28.94182
## last observed exit t = 32.21355
##
## .此時,這個數據中因這一行命令產生的新變量 _t 其實是每個實驗對象從時間點 1960/01/01 到他們各自離開實驗追蹤的時間長度。而 _t0 則是每個實驗對象從時間點 1960/01/01 到他們各自參加這個實驗的時間長度。當我們使用 strate 和 stmh 來評價高血壓和死亡之間的關係的時候會得到和之前相同的分析結果。
##
## . cd "~/Downloads/LSHTMlearningnote/backupfile/Users/chaoimacmini/Downloads/LSHTMlearningnote/backupfiles
##
## . use mortality, clear
##
## . gen hyper = .
## (4,298 missing values generated)
##
## . quietly replace hyper = 0 if systolic < 140
##
## . quietly replace hyper = 1 if systolic >= 140 & systolic != .
##
## . quietly stset exit, failure(died) enter(enter) id(id) scale(365.25)
##
## . strate hyper, per(1000)
##
## failure _d: died
## analysis time _t: exit/365.25
## enter on or after: time enter
## id: id
##
## Estimated failure rates
## Number of records = 4263
##
## +---------------------------------------------------+
## | hyper D Y Rate Lower Upper |
## |---------------------------------------------------|
## | 0 94 9.9879 9.4114 7.6888 11.5199 |
## | 1 40 1.3819 28.9456 21.2322 39.4611 |
## +---------------------------------------------------+
## Notes: Rate = D/Y = failures/person-time (per 1000).
## Lower and Upper are bounds of 95% confidence intervals.
##
##
## . stmh hyper
##
## failure _d: died
## analysis time _t: exit/365.25
## enter on or after: time enter
## id: id
##
## Mantel-Haenszel estimate of the rate ratio
## comparing hyper==1 vs. hyper==0
##
## ------------------------------------------------------------------------
## Rate ratio chi2 P>chi2 [95% Conf. Interval]
## ------------------------------------------------------------------------
## 3.076 39.30 0.0000 2.124 4.453
## ------------------------------------------------------------------------
##
## .那現在繪製的 Kaplan-Meier 生存曲線會變成什麼樣子呢?
sts graph, by(hyper)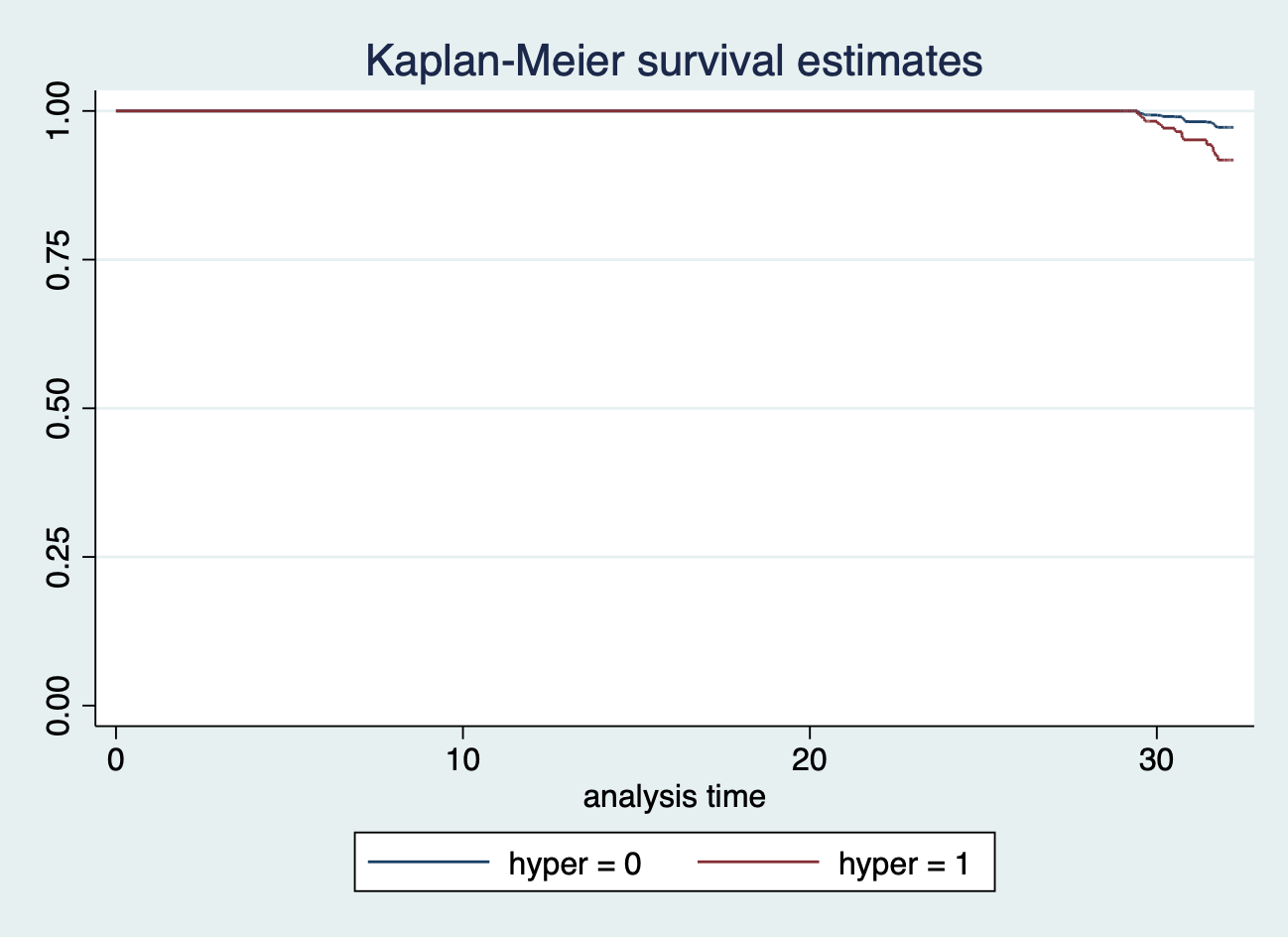
圖 117.9: sts graph, by(hyper) without specifying origin in stset command.
所以,當我們忘記設定實驗的起點時間 origin 時,Stata自動把所有實驗的時間起點設置成 1960/01/01。